
Atomistic Simulations and In Silico Mutational Profiling of Protein Stability and Binding in the SARS-CoV-2 Spike Protein Complexes with Nanobodies: Molecular Determinants of Mutational Escape Mechanisms
4.8
(634) ·
$ 4.50 ·
In stock
Description
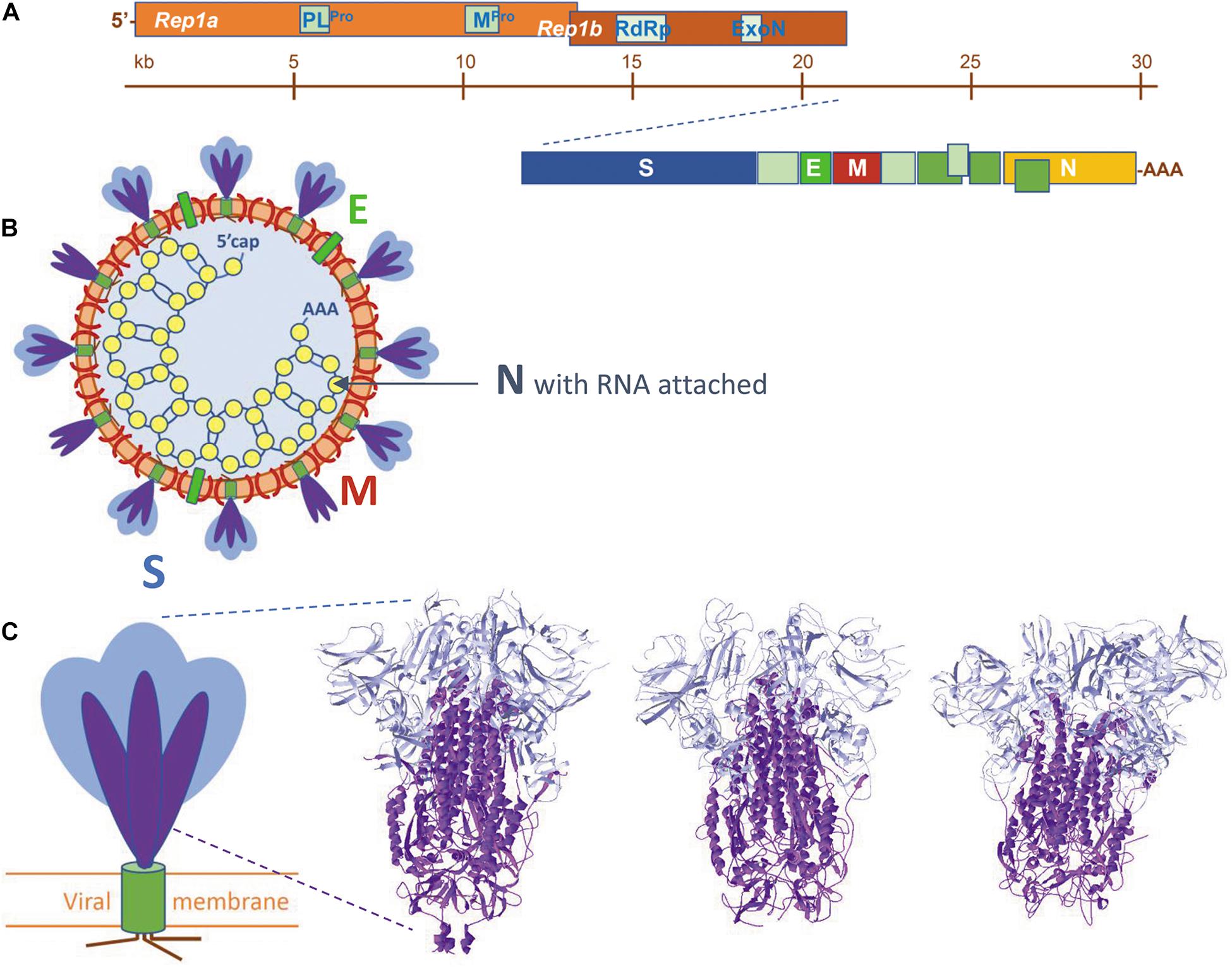
Frontiers Scrutinizing Coronaviruses Using Publicly Available Bioinformatic Tools: The Viral Structural Proteins as a Case Study

IJMS, Free Full-Text
Point mutations in SARS-CoV-2 variants induce long-range dynamical perturbations in neutralizing antibodies - Chemical Science (RSC Publishing) DOI:10.1039/D2SC00534D
In silico analysis of SARS-CoV-2 spike glycoprotein and insights into antibody binding

In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin

Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2

E484K and N501Y SARS-CoV 2 spike mutants Increase ACE2 recognition but reduce affinity for neutralizing antibody - ScienceDirect

Mutational landscape and in silico structure models of SARS-CoV-2 spike receptor binding domain reveal key molecular determinants for virus-host interaction, BMC Molecular and Cell Biology
Structural and non-structural proteins in SARS-CoV-2: potential aspects to COVID-19 treatment or prevention of progression of related diseases, Cell Communication and Signaling

In-Silico Design of a Novel Tridecapeptide Targeting Spike Protein of SARS- CoV-2 Variants of Concern
Related products
You may also like
copyright © 2019-2024 choiceworldjewellery.com all rights reserved.









